Rotavirus - Classification, Morphology, Genome, Replication, Pathogenesis, Clinical syndrome, Lab diagnosis
Classification of Rotavirus
The classification of Rotavirus is as follows:
Realm: Riboviria
Kingdom: Orthornavirae
Phylum: Duplornaviricota
Class: Resentoviricetes
Order: Reovirales
Family: Reoviridae
Subfamily: Sedoreovirinae
Genus: Rotavirus
The genus has nine species-
Rotavirus A
Rotavirus B
Rotavirus C
Rotavirus D
Rotavirus F
Rotavirus G
Rotavirus H
Rotavirus I
Rotavirus J
Characteristics of Rotavirus
Rotavirus is a major cause of acute gastroenteritis (diarrhea) in human infants (6 months to 2 years). Adults may also be infected. Serotypes A, B, and C are associated with human infections and within rotavirus A, multiple genotypes are present.
It also infects young animals.
Morphology of Rotavirus
Morphologically, Rotavirus virion measures 75nm in diameter and is icosahedral in shape, and is non-enveloped with three layered capsids. It has the characteristic of a wheel-shaped appearance which is a diagnostic feature.
The genome is double-stranded RNA and linear with 10-12 segments of the genome. The genome size is 16-27kbp. The virus also contains 60 spikes on the surface that consists of trimers of VP4.
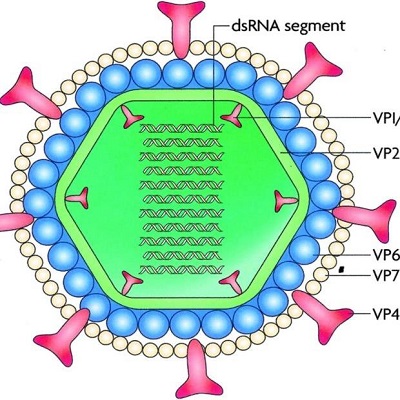
Fig: Rotavirus morphology (Source: ResearhGate)
Genome of Rotavirus
Each genome segment of Rotavirus codes for only one virus-specific protein- 6 Structural proteins and 5-6 non-structural proteins.
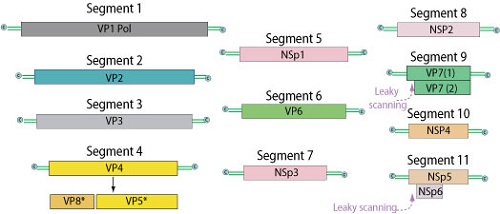
Fig: Rotavirus genome (Source: ViralZone)
Viral Protein | Function | ds RNA segment no. |
|---|---|---|
VP1 | (RNA-dependent RNA polymerase) ds RNA synthesis | 1 |
VP2 | Inner shell protein | 2 |
VP3 | Capping enzyme | 3 |
VP4 (cleaved to VP5 and VP8) | Viral attachment; determines P serotype | 4 |
VP7 | Determines G serotype | 9 |
VP6 | Middle later (shell/capsid) protein interacts with VP2 (inner layer) and VP$ and VP& (outer layer protein) | 6 |
NSP1 | IFN antagonist (degrades IFN regulatory factor IRF3 and IRF 7) | 5 |
NSP2 | Viroplasm formation | 8 |
NSP3 | Functions as polyadenylate acid-binding protein and binds to cellular eIF4G-causes systemic spread | 7 |
NSP4 | Outer capsid assembly, enterotoxin | 10 |
NSP5 and NSP6 | Viroplasm formation | 11 |
Replication of Rotavirus
Rotavirus replication occurs in the cytoplasm of infected cells. The attachment occurs via protein component VP4 to sialic acid, α2β1, HSP70.
The viral protein-receptor interaction leads to a conformational change in the viral capsid (VP4). The conformational change is brought about by the proteolytic cleavage of VP4 into VP5 and VP8. VP5 and VP8 are additional binding sites.
Rotavirus can now enter the host cell via receptor-mediated endocytosis. Passage of virion through the host cell membrane removes the outer capsid forming subviral particles. The virus never completely uncoats and viral genome RNA remains inside the sub-viral particle.
RNA-dependent RNA polymerase transcribes genomic RNA to mRNA. mRNA is 5’cap but is never polyadenylated. The viral mRNA leaves the core via channels of sub-viral particles. mRNA translates to generate viral protein. mRNA also acts as a template for RNA replication via the aid of RNA polymerase and is packed into the new capsid.
The new double-layered particle binds to NSP4 (intracellular viral receptor) and passes through ER, gaining an envelope transiently. At the same time, VP4 and VP7 are incorporated into the virion, making it mature. It then loses the envelope.
Rotavirus then pass through the Golgi apparatus from ER. Finally, virions are released from enterocytes.
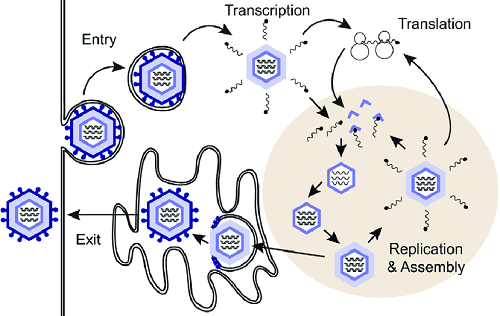
Fig: Rotavirus replication (Source: ResearhGate)
Pathogenesis of Rotavirus
Transmission occurs by the fecal-oral route. Rotavirus survives an acidic environment in the stomach and initiates infection in mucosal cells of the small intestine (does not infect the mucosa of the stomach and large intestine).
They adhere to and replicate exclusively in the differentiated epithelial cells at the tips of small intestine villi. Crypts that contain undifferentiated enterocyte stem cells are not infected.
Diarrhea includes both malabsorption and secretory component. Malabsorption of nutrients, electrolytes, and water is due to damaged mature absorptive enterocytes. The Rotavirus-induced down-regulation and expression of absorptive enzymes also take place.
The functional changes in the tight junction between enterocytes lead to paracellular leakage. Also, NSP4 is an enterotoxin that activates cellular Cl- (Chlorine ion) channels leading to increased Cl- and consequently water secretion. Neural activators are also released.
NSP1 degrades IFN regulatory factors (IR3 and IRF7)
Rotavirus-specific IgA on the enteric mucosal surface provides protection against Rotavirus
Clinical symptoms/syndrome of Rotavirus
Rotavirus causes an abrupt onset of diarrhea after an incubation period of 48 hours. Other clinical symptoms/syndromes include:
fever, vomiting, dehydration, and watery diarrhea
Vomiting is of short duration and can occur before or after the onset of diarrhea
Diarrhea stool may be watery, green, or yellow but does not contain mucus
Loss of electrolytes if not replaced, dehydration occurs and severity ranges from mild to life-threatening
Viral excretion in the stool may persist up to 50 days after the onset of diarrhea
In immunodeficient individuals, persistent severe chronic diarrhea occurs
Laboratory diagnosis of Rotavirus
The laboratory diagnosis of Rotavirus begins with the collection of samples.
Sample
stool
Microscopy
The wheel appearance of the Rotavirus particles is seen during electron microscopy.
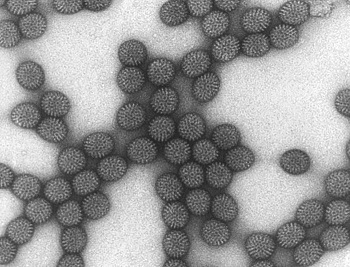
Fig: Rotavirus electron microscopy (Source: CDC)
Ag Detection
The antigen detection tests for Rotavirus include:
Enzyme-linked immunosorbent assay
Immunochromatographic assay
these Target VP6 (middle-layer protein)
Isolation
Rotavirus is difficult to grow in cell culture thus not routinely used
The addition of trypsin (proteolytic enzyme) in tissue culture medium facilitates rotavirus growth
It also cleaves outer capsid protein thus facilitating uncoating
Molecular methods
PCR (genotyping is also done via PCR) is done for Rotavirus.